Have you ever thought about how complex you are! Compare with the ones around you. You are different in all aspects. Your creativity, attitude, taste, goal, habits, phenotype may not be found anywhere else. But how precisely you have understood the bricks which create this wonder along the pathway of genomics. Yes, That is all about genome complexity.
Being astounding creatures, this complexity, and uniqueness is created by the thousands of different traits encoded by our genome. Genome, the hereditary basis of every living organism contains the biological information needed to construct and maintain a living example of that organism. The genome may consist of DNA or RNA. As few viruses only account for RNA genomes, here the attention is mainly paid to the DNA genomes. The most significant part of the nuclear genome of Eukaryotes is responsible for the bulk of the genome in the chromosomes of the cell. To provide a highly packed structure, they have arranged respectively 10nm chromatin fiber, 30 nm solenoid model, 300nm loops and 250nm compressed fiber arrangement.
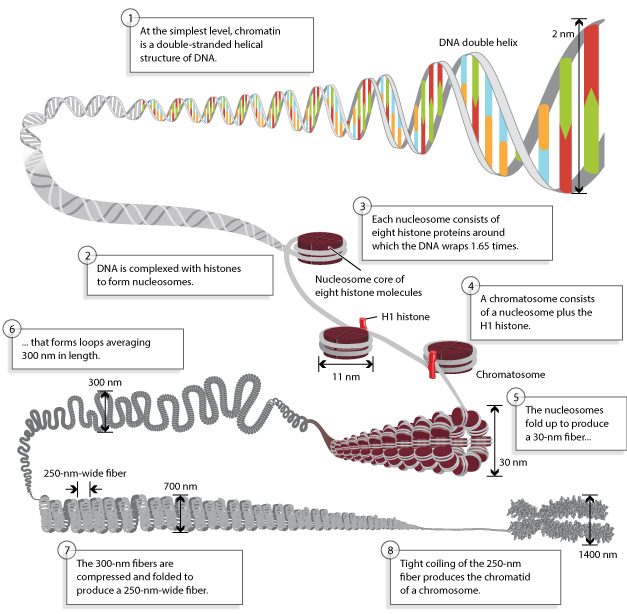
Outlining the complexity, several molecular characterization techniques like whole genome sequencing and DNA reassociation kinetics have shown that genome complexity is a result of several types of DNA groups present in the eukaryotic genome. Anyhow, the chromosome number, in a haploid cell does not show any relationship to the complexity of the organism. As C- value paradox describes C value is poorly correlated with the complexity of the genome.

So what makes us this complex? Those are the several types of coding, non-coding, repeated, non – repeated, moderately repeated genome regions. As mentioned, DNA reassociation kinetics have identified three major classes of DNA sequences in eukaryotic genomes based on the frequencies they found in the genome.
- Highly repeated fraction (highly repetitive sequences) consists of > 105copies per genome
- Moderately repeated fraction (moderately repetitive sequences) consists of a few to 105copies per genome
- Non-repeated fraction (unique sequences) occurring only one copy per genome
Here, the non-repeated DNA fraction is mainly due to the single copy of DNA or gene families. A single copy of DNA is assumed to be present in single or very low copy numbers and may contain protein-coding genes but at some points, they can be spacer DNA, intergenic spacer sequences, or intronic sequences.
In multicellular organisms, 25-50% of protein coding genes are single copy genes. In eukaryotes that DNA fraction is expected to contain most of the genes encoding mRNAs, mostly the genes exhibit a Mendelian pattern of inheritance.
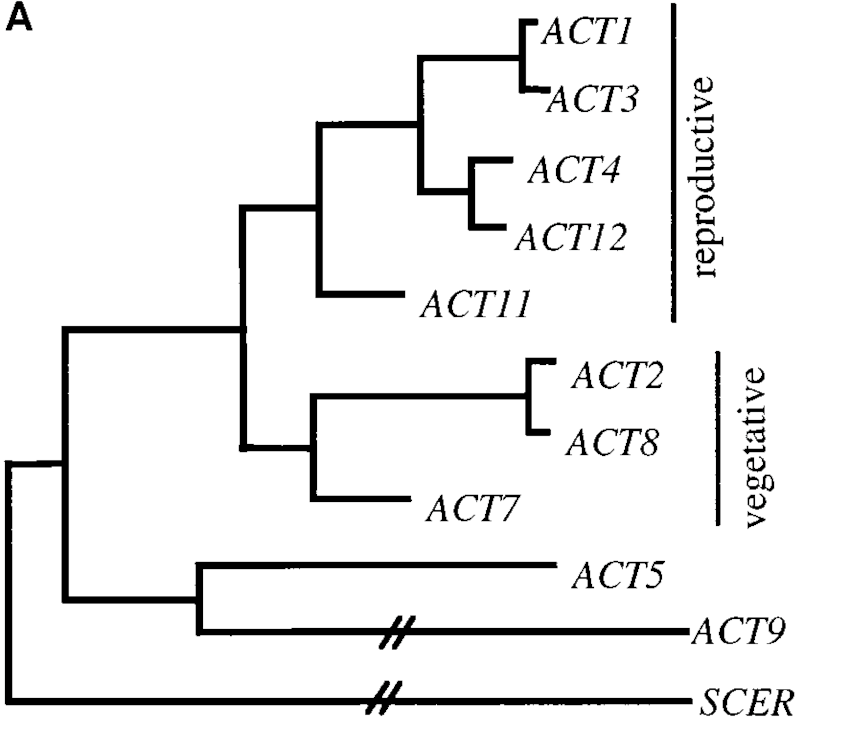
The remaining protein-coding non-repeated genes belong to gene families signifying one factor that contributes to the Genome Complexity. Being a set of several similar genes formed by duplication of a single original gene, a gene family generally has similar biochemical functions. Different members of a gene family may be transcribed in different tissues or at different developmental stages. For example family of genes for human hemoglobin subunits and Arabidopsis Actin gene family can be identified.
The human β-like globin gene family encodes β-subunits of hemoglobin which contains five functional genes on the chromosome 11 — β, δ, Aγ, Gγ, and ϵ. showing tandem arrangement. During human development, genes are expressed at different times and in different tissues. Hemoglobins containing either Gγ or A γ are expressed only during fetal life.

The actin (ACT) gene family in plants regulates many different cellular processes like cell elongation, tip growth of pollen tubes. It contains 10 genes on four different chromosomes, chromosomes 1, 2, 3, and 5 . Thus, 08 functional genes and 02 pseudogenes are there, showing the dispersed arrangement.
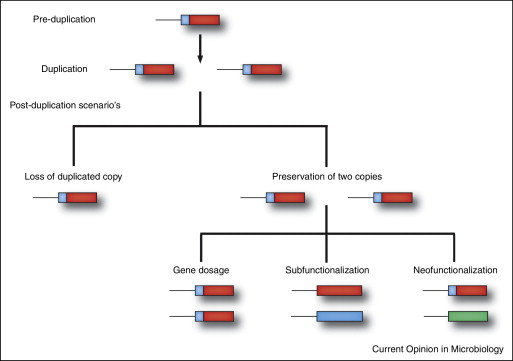
In evolution duplication of an original ancestral gene may result, gene families. Thus, duplicated genes then diverge as a consequence of the accumulation of mutations during evolution. Gene duplication may occur due to Whole genome duplication, Tandem duplication, or Retroduplication. Whole genome duplication (polyploidization) occurs through an increase in ploidy which means having more than the usual two homologous sets of chromosomes. As an extreme mechanism of gene duplication that results in a sudden increase in both genome size and the entire gene set. Along the journey of evolution, WGDs that have occurred in the lineages of several domesticated crop species (ex- wheat, cotton, soybean) and have contributed to important agronomic traits showing that WGD accounts for the majority of duplicated genes especially in plants.
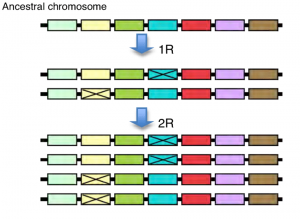
Tandem duplication generates clusters of two to many paralogous sequences with no or few intervening sequences. Therefore duplicated genes are found adjacent to the original gene in a chromosome. This results due to the unequal crossing-over events that occur via misalignment of homologous chromosomes during meiosis. The position of crossing-over determines whether the duplicated region will remain part of a gene, an entire gene, or several genes.

In retroduplication, processed mRNA is reverse transcribed to DNA (cDNA) and then inserted into the genome more or less random way as dispersed duplicates, resulting in a lack of regulatory elements like introns and other regulatory sequences for transcription.
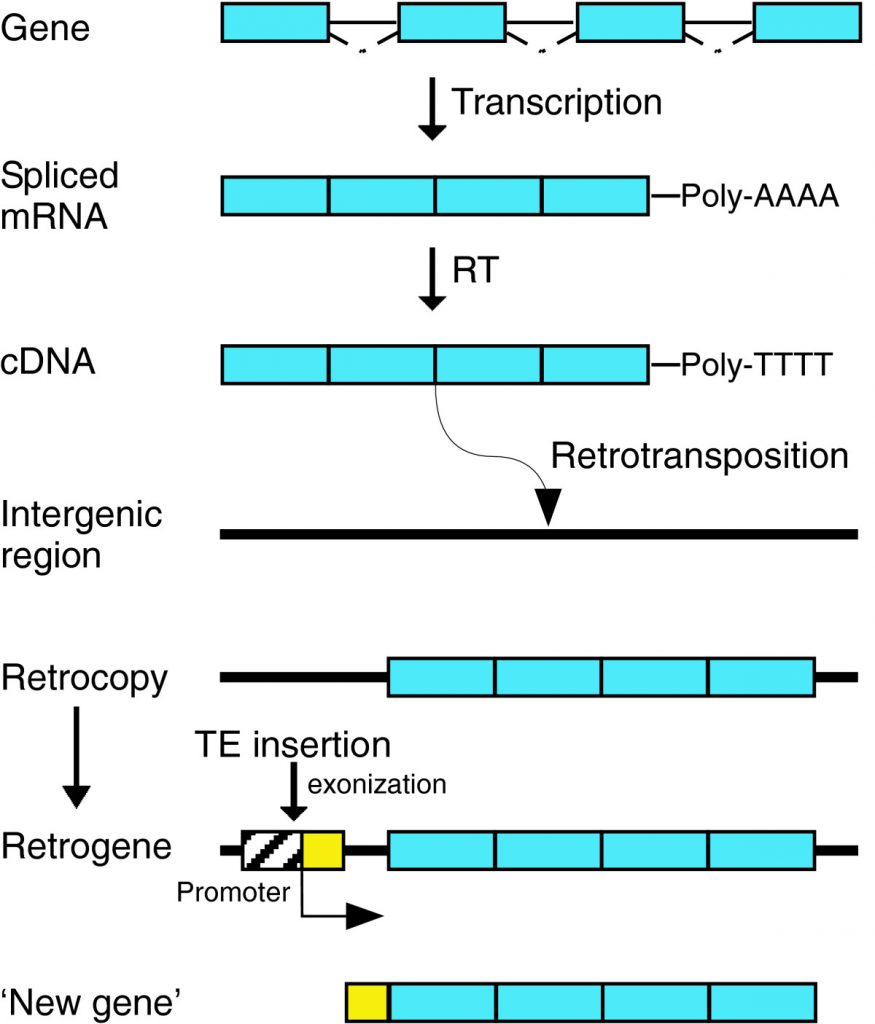
Resulted fates may be retained without acquiring new functions (Dosage balance, Gene dosage increase, Sub-functionalization) or acquiring new functions. (Neo functionalization).
In gene dosage increase both duplicates are retained because of a beneficial increase in expression resulting in increased metabolic activity. Sub-functionalized duplicates can be stably maintained when they differ in some aspects of their function, and randomly losing sub-functions at the level of protein function or gene expression or at both levels simultaneously. Thus no novel functions are evolved, only partitioning of the ancestral functions happens.
In dosage balance, duplicate genes that are dosage-sensitive tend to be retained to maintain balance causing a gene with a great number of molecular interactions to be duplicated with its interactors, retention of both copies is favorable because just removal of one gene would lead to imbalance. Neo functionalized both duplicates are retained because of a gain-of-function after duplication contributing to better fitness for the trait.
Therefore the complexity of the genome is a vital factor that represents the structures of coding or non-coding units for maintaining life. Thus here we highlighted how non-repetitive DNA accounts for the well-known, genome complexity. But there are many a thousand facts to be discussed related to this particular scenario. Let us walk along the path of genomics through another episode as well.
Image Courtesies:
- Featured Image – https://bit.ly/3lT7oNO
- Figure 1 – https://bit.ly/3zKtu9Y
- Figure 2 – https://bit.ly/3AIC5Ly
- Figure 3 – https://bit.ly/3i6TbeX
- Figure 4 – https://bit.ly/3zDYo3Q
- Figure 5 – https://bit.ly/3AEgT9y
- Figure 6 – https://bit.ly/3m0oQQx
- Figure 7 – https://bit.ly/3kDxt3R
- Figure 8 – https://bit.ly/3EM22wh
References:
- https://journals.plos.org/
ploscompbiol/article?id=10. 1371/journal.pcbi.1008222 - https://www.science.org/doi/
abs/10.1126/science.1089370 - Molecular Biology of the Cell (6th Edition) by Bruce Alberts, Alexander Johnson, Julian Lewis, David Morgan, Martin Raff, Keith Roberts, and Peter Walter.

