The influenza pandemic of 1918-19, or more commonly known as the Spanish Flu was a global tragedy; one of the worst ever to befall humanity. It infected an estimated 500 million people worldwide, which was about one-third of the world population at that time, in about just 18 months and killed an estimated 20 to 50 million victims. The Spanish flu was first observed in Europe, the United States, and part of Asia before swiftly spreading around the world with astonishing speed, overwhelming India and reaching Australia and remote Pacific islands.

Influenza or flu is caused by a virus that attacks the respiratory system. The flu virus is highly contagious and is transmitted from person to person through airborne respiratory secretions. That is when an infected person coughs, sneezes, or talks, respiratory droplets are generated and transmitted into the air and can be inhaled by anyone nearby. Besides, if people touch something with the virus on it and then touch their nose, mouth, or eyes, they can be easily infected. Under such circumstances, if a new strain of influenza virus emerges against which the population has no immunity, an outbreak can occur. Thus a flu pandemic such as in 1918 occurs when an especially virulent new influenza strain for which it is little or no immunity appears and spreads quickly from one person to another around the globe (History.com Editors, 2010).
A strain can be simply explained as a genetic variant or a subtype of a particular microorganism that can be differentiated using the genetic makeup. When one such type undergoes either subtle or major genetic changes through mutations or recombinations respectively, a new strain is said to be created and their phenotypic expressions like antigens on them, change accordingly.
If a pathogenic strain infects the human body, it develops immunity against that particular strain by creating antibodies. This immunity usually prevails for a considerable time before degrading. However, if a new strain enters the body immediately since they are genetically and antigenically different, antibodies already present can’t recognize the new strain and is useless against it. So the body is said to be lack of immunity for the new strain.
Any of several closely related viruses in the family Orthomyxoviridae (a group of RNA viruses) can cause influenza. They are categorized as types A, B, C, and D which generally produce similar symptoms. However, they are completely unrelated antigenically. Hence an infection with one type confers no immunity against the others obviously when the body can’t confer immunity even against different strains of the same type. Type A causes the great influenza epidemics and are the most lethal ones, and the B viruses cause smaller localized outbreaks. Type C causes only mild respiratory illness. Influenza D is not known to infect humans and has been observed only in pigs and cattle.
Being an Orthomyxovirus, the influenza virus is enveloped with the characteristic spike-like viral glycoproteins called Haemagglutinin and Neuraminidase. Influenza A viruses are classified into subtypes and are further divided into strains. Based on two surface antigens — hemagglutinin (H) and neuraminidase (N), subtypes of influenza A are differentiated. Thus their description of H1N1, H5N1, etc. are relying on the type of H or N antigens.
Influenza is very common among people and causes seasonal flu generally running from late fall into spring. However especially virulent new strains of influenza A virus has unfortunately been responsible for three global pandemics in the last century: the Spanish Flu in 1918, Asian Flu in 1957, and the Hong Kong Flu in 1968. The influenza pandemic 1918-19 resulted from an influenza virus called “influenza type A subtype H1N1”. It was not until the 1930s that an H1 subtype was identified as the causative strain for which initially attributed to Haemophilus influenzae. Over the years, studies of individual genes and the fully reconstructed live 1918 virus have identified numerous features that may have likely contributed to its robustness and rapid global spread.
It is accepted that animals including pigs, horses, and wild birds are a natural reservoir for influenza A viruses. Yet, with vast antigenic and genetic diversity inherent among the influenza virus and their surface proteins, a strain to which humans are immunologically naïve can jump the species barrier easily. A-(H5N1) and A-(H7N9) viruses are two such examples (Belser & Tumpey, 2018).
A zoonosis is a disease or infection that normally exists in vertebrate animals and is naturally transmissible to humans. Hence animals play an essential role in maintaining zoonotic infections in nature. Zoonoses may be bacterial, viral, or parasitic, or may involve unconventional agents. The ability to infect humans may come as a result of one of three plausible scenarios. First is the selection within animal host before zoonotic transfer where a precursor pathogen undergoes mutation to acquire the ability to bind to human cells and infect them. The second case is the selection during the passage from host animal to human may be via an insect vector. The third scenario is natural selection within humans following the zoonotic transfer meaning the pathogen gets mutated within the human body after being transferred and starts a pandemic. (Andersen, et al., 2020)
The influenza virus is infamous for its ability to mutate where a new strain appears to be a result of reassortment of human influenza and swine influenza viruses, in all four different strains of subtype H1N1. Mixing of the genes of one species so that a new combination arise within different individuals can be stated as reassortment. This process has been responsible for some of the major genetic shifts experienced in pandemics. This strain of the H1N1 Influenza A subtype is a result of a mixing of the virus from humans, birds, and pigs (Lim & Mahmood, 2011).
Newly surfaced influenza viruses tend to be initially highly infectious and virulent in humans. This is because they possess novel antigens to which the human body has no prepared immune defense or existing antibodies. However, the infectiousness and virulence of the virus decrease once a significant proportion of a population develops immunity through the production of antibodies capable of neutralizing the new virus. This is called ‘herd immunity’. That means, once the majority of a population develops immunity against infection, it provides an indirect form of protection to those that are not immune to that particular infection. This is achieved by the prevention of the infection further by the immune portion of the population.
Clinical features of Spanish flu usually included sudden fever and cough. These were typically associated with at least one of the other symptoms of chills, lethargy, dehydration, headache, sore throat, coryza, diarrhea, vomiting, abdominal pain, myalgia, or arthralgia. Patients becoming short of breath and lack of enough oxygen made their faces appeared tinged with blue. Hemorrhages filled the lungs with blood and vomiting and nosebleeds made victims drown in their body fluids.
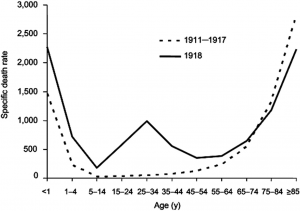
Usually, outbreaks of influenza viruses are generally most fatal to young children and the elderly. Yet, Spanish flu being an exceptional case, the fatality rate in people between ages 20 and 40 became unexpectedly high. This phenomenon identified as a result of ‘Cytokine storm’ is now believed to be due to hyper-reaction of the immune system of physically fit youth to new strains of the influenza virus. That is overproduction and release of inflammatory substances called cytokines on an excessive scale lead to severe inflammation. In contrast, individuals whose immune systems are not fully developed (such as infants) or are weakened (such as the elderly) cannot generate such a lethal immune response.
The Spanish flu pandemic occurred in 3 waves. The 1st originated in early March 1918, during World War I. The virus quickly spread through Western Europe even though its birthplace remains unclear. By July it had spread to Poland. Although the 1st wave of influenza was comparatively mild, during the summer a lethal type of disease was recognized as a 2nd wave. This form fully emerged in August 1918. It was often observed that Pneumonia developed quickly, with death following two days after the 1st indications of the flu were detected. The 3rd wave of pandemic hit during the following winter, and by spring the virus had run its course. In the two later waves, surprisingly about half the deaths were among 20 to 40-year-olds. This was a remarkable deviation from mortality age patterns for influenza (The Editors of Encyclopaedia Britannica, 2020).
To put the global mortality burden for the 1918 pandemic in perspective, one can compare it with the other influenza pandemics. Implementing this technique shows the 1918 burden was very high. Apart from those other aspects of the influenza pandemics were quite similar. However, when estimating the number of deaths, it is important to test the theoretical feasibility of estimates, because the results of such tests suggest the true estimate is very unlikely to be higher than 25 million and probably closer to a final estimate of 17.4 million deaths. (Spreeuwenberg, et al., 2018).
The Spanish flu took a heavy human toll. It wiped out entire families and left countless widows and orphans in its wake. Bodies kept piling up and funeral parlors were overwhelmed. Many people had to dig graves for their family members. Unfortunately, the flu was also detrimental to the economy. Businesses were forced to shut down as so many employees were sick. Even most basic services like mail delivery and garbage collection were interrupted due to sick workers.
Sometimes there weren’t enough farm workers to harvest crops. Even state and local health departments had to be closed for business. This hampered the efforts to chronicle the spread of the pandemic and provide the public with answers about it. Without any kind of vaccine for protection and effective antibiotics to tackle secondary bacterial infections that followed influenza infections, control efforts worldwide were restricted to non-pharmaceutical interventions. The use of antiseptics and disinfectants, isolation, quarantine, practicing good personal hygiene, and limitations of public gatherings, were such measures.

To worsen the matters, unfortunate incidents of Aspirin poison occurred. Many doctors, at the mercy of having no cure for the flu, prescribed medications that they thought would alleviate symptoms. These medications included Aspirin advised to take up to 30 grams per day, a dose which is now known to be toxic. Symptoms of Aspirin poisoning included hyperventilation and pulmonary edema. This is now believed to have hastened many of the deaths (History.com Editors, 2010).
Knowing all this information, the question arises that what is our readiness today? International health agencies and research laboratories work in collaboration. They try to track influenza virus evolution, evaluate antigenic drift among circulating and vaccine strains, and sequence viral genes with the hope of advanced surveillance and preparedness. Production of improved vaccines along with diagnostic tools and better access to therapeutic agents with no doubt represent resources that were not available a century ago.
Yet, the novel corona pandemic of 2020 is spreading around the world putting a trail of bodies in its path. As countries race to find a cure for COVID 19 and people being arrested to one place in an attempt to prevent spreading the disease, it makes everyone wonder will the development of science and technology ever be sufficient?
Philosopher George Santayana pointed out, “Those who cannot remember the past are condemned to repeat it”. We are no doubt more prepared in 2020 for an infectious disease threat than in 1918. But under current circumstances, it is critical to remember that preparation only stems from a global commitment to share data about viral isolates, support innovative research, and dedicate resources to predict the pandemic risk of new influenza viruses arising from zoonotic reservoirs.
Image Courtesies
- Featured image: http://t.ly/EDXL
- 1st content image: https://bit.ly/2YYSn31
- 2nd content image: https://bit.ly/2YZSrzF
- 3rd content image: https://bit.ly/3bsMHkr
- 4th content image: http://t.ly/hcEd
- 5th content image: http://t.ly/NnPJ
References
Andersen, K. G. et al., 2020. The proximal origin of SARS-CoV-2. Nature Medicine, 17 March, Issue 26, pp. 450-452.
Belser, J. A. & Tumpey, T. M., 2018. The 1918 flu, 100 years later. Science, 19 January, 359(6373), p. 255.
History.com Editors, 2010. HISTORY. [Online]
Available at: https://www.history.com/topics/world-war-i/1918-flu-pandemic
[Accessed 30 April 2020].
Lim, B. H. & Mahmood, T. A., 2011. Influenza A H1N1 2009 (Swine Flu) and Pregnancy. Journal of Obstetrics & Gynecology of India, 23 September, 4(64), pp. 386-393.
Spreeuwenberg, P., Kroneman, M. & Paget, J., 2018. Reassessing the Global Mortality Burden of the 1918 Influenza Pandemic. American Journal of Epidemiology, 7 September, 187(12), pp. 2561-2567.
The Editors of Encyclopaedia Britannica, 2020. The influenza pandemic of 1918–19. [Online]
Available at: https://www.britannica.com/event/influenza-pandemic-of-1918-1919
[Accessed 30 April 2020].
Reviewed by: Mr. Pubudu Nawarathna (from McGill University / Canadian Center for Computational Genomics, Canada – Specialized in Biology (Plant sciences and Zoology), Computer Science and Information Technology, Molecular Biology)

