“It took a great deal of work on both sides of the Atlantic, but it was worth the effort – we were delighted to see that our new chimeric enzyme is up to three times faster than the naturally evolved separate enzymes, opening new avenues for further improvements.”
-Professor John McGeehan, University of Portsmouth-
In the modern world, plastics have become a major global crisis that has negatively impacted the environment severely, even more than we could assume. Around one million plastic bottles are being purchased every minute around the world and more dangerously, about 50 percent of them are designed for single-time usage. Due to the rapid accumulation of plastics in the marine ecosystems, a new marine microbial habitat known as the “plastisphere” has been introduced to the science world too. Although many actions such as recycling, minimal usage of plastics and reusing are being implemented, they are not effective to minimize this crisis.

Polyethylene terephthalate (PET) is a polymer that is found in most consumer packaging including cookie containers, soda bottles, packaging of fruits, and certain fibers and textiles. After years of research, in 2018, scientists discovered an enzyme known as PETase(PET hydrolase), a plastic-eating enzyme that breaks down polyethylene terephthalate (PET). It was fascinating to discover that this enzyme could complete a full circular process by both depolymerizing and repolymerizing. However, with the intention to increase the efficiency of plastic degradation, PETase was modified to build up a new enzyme known as FAST-PETase (functional, active, stable and tolerant PETase). This discovery was quite a remarkable event in the science world as the scientists were intelligent enough to discover an enzyme that operates efficiently at low temperatures making it affordable and portable at an industrial scale.
The enzyme was synthesized by a type of bacteria known as Ideonella sakaiensis, which feeds on plastic bottles. This, in fact, is an outstanding solution to the daily increasing plastic crisis. Ideonella sakaiensis is a bacterium that belongs to the genus Ideonella and family Comamonadaceae, possessing the ability to break down PET for carbon and energy usage. This bacterium usually takes up to 60 weeks to degrade a wafer-thin plastic in the laboratory. As the time taken for the degradation is high, a super enzyme known as MHETase was revealed making the depolymerization even more efficient. Yet scientists carried out their research further to discover more potent processes and enzymes. This resulted in the invention of a hybrid enzyme known as “cocktail” produced by combining PETase and MHETase, which could degrade PET six times faster than the original enzyme.
History
In 2016, the discovery of the soil bacterium, Ideonella sakaiensis was reported in Japan, found growing in PET contaminated plastic. This bacterium is a gram-negative, aerobic, rod-shaped bacterium that uses a two enzyme system to degrade PET.

What is an enzyme?
Enzymes are proteins that act as catalysts regulating the rate of chemical reactions, without being used up. Enzymes play a major role in regulating most biological processes, altering the energy barrier. The enzyme activity depends on many factors such as temperature, pH, substrate concentration, etc. They react with the substrate to form an intermediate complex which is the transition state reducing the amount of energy needed to overcome the energy barrier.
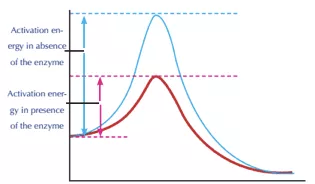
Mechanism of the plastic degrading enzyme
As shown in the above graph, the reaction mechanism takes place in two stages, acylation and deacylation. Each step is a single, associative, concerted step. Acylation consists of a proton transfer associated with a nucleophilic attack on the substrate. This leads to a tetrahedral transition state, which results in the release of the MHET after breaking the ester bond. Deacylation occurs by deprotonation of an active site of water molecule followed by the resulting hydroxide attacking the intermediate and breaking its bonds to the substrate. Subsequently, enzyme regeneration occurs. The energy barrier of the rate-limiting acylation is 20 kcal mol-1. This barrier is reduced by the mutation of a few compounds. Finally, the PET is broken down into its monomers along with the regeneration of the enzyme.
As the thermoplastic polymer PET could be degraded to its monomers with the aid of “cocktail”, the degradation could be done completely within a few hours. Therefore, these monomers could be reassembled to form new utensils, making reuse possible. However, the usage of plastic should be minimized as much as possible, by using other alternatives.
Written by: Ishana Paramee
Image courtesy:
- Featured image: https://bit.ly/3wHukFa (Customized By Umesha Abeysuriya)
- Content Image 1: https://bit.ly/3wMzL5G
- Content Image 2: https://bit.ly/3NrUhyY
- Content Image 3: https://bit.ly/3PHAxJF
- Content Image 4: https://bit.ly/3wFwp5x
References:
- (New Plastic-Eating Enzyme ‘Cocktail’ Can Digest Plastic Waste Six Times Faster, n.d.)
- (Plastic-Eating Enzyme Could Eliminate Billions of Tons of Landfill Waste, n.d.)(Yoshida et al., 2016)
- (Han et al., 2017)Han, X., Liu, W., Huang, J. W., Ma, J., Zheng, Y., Ko, T. P., Xu, L., Cheng, Y. S., Chen, C. C., & Guo, R. T. (2017). Structural insight into catalytic mechanism of PET hydrolase. Nature Communications, 8(1). https://doi.org/10.1038/S41467-017-02255-Z
- New Plastic-Eating Enzyme ‘Cocktail’ Can Digest Plastic Waste Six Times Faster. (n.d.). Retrieved May 12, 2022, from https://www.greenqueen.com.hk/new-plastic-eating-enzyme-cocktail-can-digest-plastic-waste-6-times-faster/
- Plastic-eating enzyme could eliminate billions of tons of landfill waste. (n.d.). Retrieved May 12, 2022, from https://phys.org/news/2022-04-plastic-eating-enzyme-billions-tons-landfill.html
- Yoshida, S., Hiraga, K., Takehana, T., Taniguchi, I., Yamaji, H., Maeda, Y., Toyohara, K., Miyamoto, K., Kimura, Y., & Oda, K. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science, 351(6278), 1196–1199. https://doi.org/10.1126/SCIENCE.AAD6359
- Han, X., Liu, W., Huang, J. W., Ma, J., Zheng, Y., Ko, T. P., Xu, L., Cheng, Y. S., Chen, C. C., & Guo, R. T. (2017). Structural insight into catalytic mechanism of PET hydrolase. Nature Communications, 8(1). https://doi.org/10.1038/S41467–02255-Z
- e). Science, 351(6278), 1196–1199. https://doi.org/10.1126/SCIENCE.AAD6359